2017/08/18
Direct detection of early-stage cancers using circulating tumor DNA
<Abstract>
Early detection and intervention are likely to be the most effective means for reducing morbidity and mortality of human cancer. However, development of methods for noninvasive detection of early-stage tumors has remained a challenge. We have developed an approach called targeted error correction sequencing (TEC-Seq) that allows ultrasensitive direct evaluation of sequence changes in circulating cell-free DNA using massively parallel sequencing. We have used this approach to examine 58 cancer-related genes encompassing 81 kb. Analysis of plasma from 44 healthy individuals identified genomic changes related to clonal hematopoiesis in 16% of asymptomatic individuals but no alterations in driver genes related to solid cancers. Evaluation of 200 patients with colorectal, breast, lung, or ovarian cancer detected somatic mutations in the plasma of 71, 59, 59, and 68%, respectively, of patients with stage I or II disease. Analyses of mutations in the circulation revealed high concordance with alterations in the tumors of these patients. In patients with resectable colorectal cancers, higher amounts of preoperative circulating tumor DNA were associated with disease recurrence and decreased overall survival. These analyses provide a broadly applicable approach for noninvasive detection of early-stage tumors that may be useful for screening and management of patients with cancer.
<Finding>
The detection and analysis of cell-free DNA in patients’ blood are becoming increasingly accepted in oncology. However, this approach has generally been applied for the monitoring of patients with existing tumors. It has not been useful for early diagnosis of cancer because of insufficient sensitivity to detect really small tumors that only shed minute quantities of DNA into the blood, as well as difficulties with identifying cancer-associated genetic changes without knowing what mutations are present in the primary tumor. A method developed by Phallen et al., called targeted error correction sequencing, addresses both of these limitations and demonstrates the feasibility of detecting circulating cell-free DNA from many early tumors, suggesting its potential use for cancer screening.
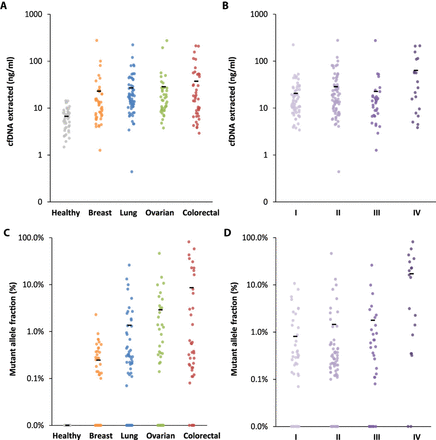
<Figure 3. cfDNA and ctDNA in healthy individuals and patients with cancer>
Amount of cfDNA extracted from all healthy individuals and patients with different cancer types (A) and from cancer patients of different stages (B). Mutant allele fraction of ctDNA detected in healthy individuals and patients with different cancer types (C) and in cancer patients of different stages (D). Means for each group are represented by the black bars in the columns analyzed. In patients for whom multiple alterations were detected, the highest value is indicated.
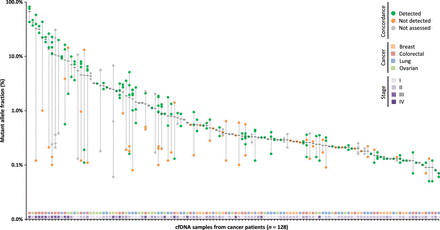
<Figure 5. Concordance between alterations in plasma and tissue>
Mutant allele fractions observed in the plasma are indicated for each alteration identified with a black bar at the mean. The presence of alterations in matched tumor specimens is indicated with green dots, whereas nonconcordant alterations are indicated in orange, and those that are not assessed are indicated in gray. Stage and cancer type for each patient are plotted in the two horizontal tracks at the bottom of the figure.