2018/01/04
The cutoff limit of reference intervals for mutant KRAS was determined to be ~ 0.09% based on samples from healthy individuals
<Title>
An improved digital polymerase chain reaction protocol to capture low‐copy KRAS mutations in plasma cell‐free DNA by resolving ‘subsampling’ issues
<Abstract>
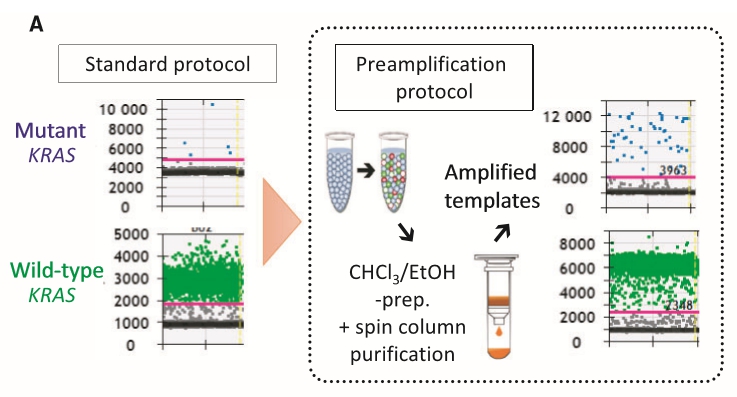
Genetic alterations responsible for the initiation of cancer may serve as immediate biomarkers for early diagnosis. Plasma levels of cell‐free DNA (cfDNA) in patients with cancer are higher than those in healthy individuals; however, the major technical challenge for the widespread implementation of cfDNAgenotyping as a diagnostic tool is the insufficient sensitivity and specificity of detecting early‐stage tumors that shed low amounts of cfDNA. To establish a protocol for ultrasensitive droplet digital polymerase chain reaction (ddPCR) for quantification of low‐frequency alleles within a limited cfDNA pool, two‐step multiplex ddPCR targeting eight clinically relevant mutant KRAS variants was examined. Plasma samples from patients with colorectal (n = 10) and pancreatic cancer (n = 9) were evaluated, and cfDNA from healthy volunteers (n = 50) was utilized to calculate reference intervals. Limited cfDNA yields in patients with resectable colorectal and pancreatic cancers did not meet the requirement for efficient capture and quantification of rate mutant alleles by ddPCR. Eight preamplification cycles followed by a second‐run ddPCR were sufficient to obtain approximately 5000–10 000 amplified copies per ng of cfDNA, resolving the subsampling issue. Furthermore, the signal‐to‐noise ratio for rare mutant alleles against the extensive background presented by the wild‐type allele was significantly enhanced. The cutoff limit of reference intervals for mutant KRAS was determined to be ~ 0.09% based on samples from healthy individuals. The modification introduced in the ddPCR protocol facilitated the quantification of low‐copy alleles carrying driver mutations, such as oncogenic KRAS, in localized and early‐stage cancers using small blood volumes, thus offering a minimally invasive modality for timely diagnosis.
<Reference>
Mol Oncol. 2017 Oct; 11(10): 1448–1458.